The Mueller-Planitz lab studies the core components of chromatin – the nucleosomes and the machinery that places them in the genome. Nucleosomes are crucial to human health. Aging, for instance, disrupts the nucleosome landscape, destabilizing the genome, and mutations in nucleosomes are drivers of cancers. Correspondingly, the locations of nucleosomes in the genome are precisely controlled by so called nucleosome remodeling complexes.
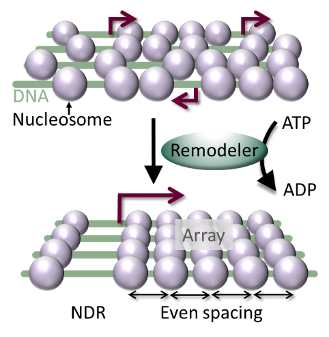
Figure: Remodeling enzymes establish the nucleosome organization over genes. In an ATP hydrolysis-dependent manner, remodelers position the first nucleosome downstream of the transcription start site (red arrow; bottom), and induce even spacing between nucleosomes downstream of it. Without remodelers (top), cryptic promoters open, leading to spurious transcription.
Aim:
The overarching aim of the Mueller-Planitz lab is to elucidate the biogenesis of the nucleosome landscape and dissect its biological function under physiological and pathological conditions. To achieve this goal, his lab bridges methodologies of molecular biology, genetics, genomics, biophysics, structural biology, and enzymology. They develop cutting-edge technology to visualize individual nucleosome patterns in single cells, and to dissect the mechanism of nucleosome remodelling genome-wide in vivo and in vitro.
Nucleosomes have recently been shown to phase separate, raising a host of fundamental biological questions. For instance, can enzymes that operate on DNA and nucleosomes freely enter chromatin condensates, or are they excluded? (How) can enzymes operate in chromatin condensates? Are they differently regulated in such an environment? Can their activity dissolve condensates or induce their formation? The Mueller-Planitz lab is addressing these and similar questions using high-end microscopy, incl. FLIM-FRET, quantitative biochemistry and computational simulations.
The Team:
Felix Müller-Planitz
Technische Universität Dresden
Institute of Physiological Chemistry
Fiedlerstraße 42
01307 Dresden, DE

