Background:
Subcellular organization via biomolecular condensation in prokaryotes has recently gained momentum with the demonstration that the fundamental process of ribosomal RNA (rRNA) transcription takes place within condensates, similar to the eukaryotic nucleolus. Specifically, NusA, an essential transcription anti-termination factor, was reported to undergo liquid-liquid phase separation (LLPS) in E. coli and to form the basis of transcription condensates in fast growing cells (Ladouceur & Weber, PNAS, 2020). However, how NusA drives LLPS and how condensates relate to transcriptional efficiency are yet to be described. Our own work has also recently shown that NusA sits at the nexus of transcription-translation coupling in the genome-reduced pathogen Mycoplasma pneumoniae (O’Reilly, Xue, …, Mahamid, Rappsilber, Science, 2020). The M. pneumoniae NusA has evolved a highly disordered C-terminal domain, which substitutes the structured C-terminal domains in E. coli and is completely absent in B. subtilis. We show that M. pneumoniae NusA also phase separates in vitro and following heterologous expression in E. coli cells.
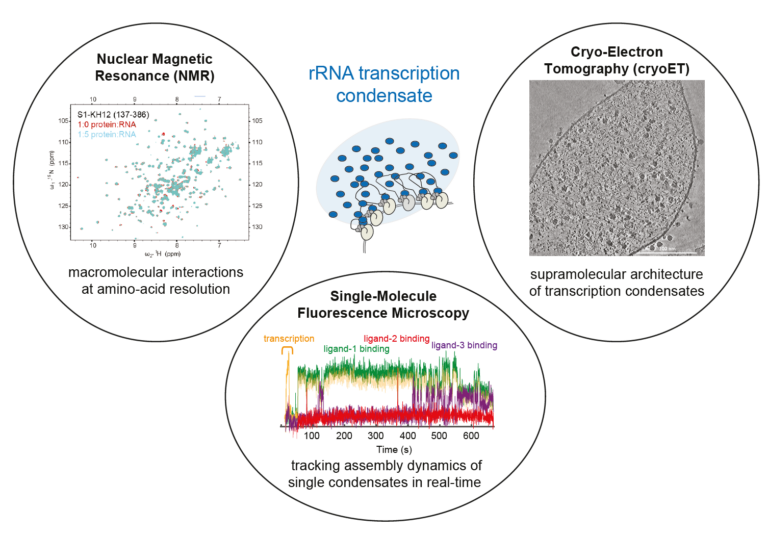
Integrative and multifaceted approach to study structure and dynamics of rRNA transcription condensates in bacteria.
Aim:
In this proposal, we aim to take advantage of the sequence hypervariability among NusA homologues across the three different bacteria to dissect the principles of phase separation, mechanisms of condensate nucleation, growth dynamics, and supramolecular structure and function in the context of the NusA-containing rRNA transcription antitermination complex. We will combine biochemical and functional assays, structural studies across scales using nuclear magnetic resonance spectroscopy and a novel combination of single-molecule fluorescence microscopy and cryo-electron tomography to provide a mechanistic and quantitative understanding of transcription condensates. Overall, we will provide the community with novel approaches in integrative modeling of molecular structure and dynamics in condensates.
The Team:

Olivier Duss
PhD
EMBL Heidelberg
Meyerhofstrasse 1
69117 Heidelberg, DE
Co-worker:
Benjamin Lau
About us
The Duss lab combines multi-color single-molecule fluorescence microscopy, structural biology and biochemistry to understand how protein-RNA complexes assemble in various context.

Julia Mahamid
PhD
EMBL Heidelberg
Meyerhofstrasse 1
69117 Heidelberg, DE
Co-worker:
Benjamin Lau
About us
The Mahamid group develops and employs advanced cryo-electron microscopy methods for in-cell structural biology to elucidate the structural principles and cytoplasmic environment driving the dynamic assembly of phase-separated compartments.

Janosch Hennig
PhD
University of Bayreuth
Universitätsstrasse 30
95447 Bayreuth, DE
Co-worker:
Tamara Bosnjakovic
About us
The Hennig group performs integrative structure modelling of RNA-protein complexes involved in transcription and translation by combining data derived mainly from nuclear magnetic resonance spectroscopy, X-ray crystallography, small-angle scattering and cryo-electron microscopy.

